Last modified: Mon May 2, 2012 11:25AM
Intro
The SimDAL specification is intended to ease the access to the information on experiments and their results as well as to reduce the required effort from the provider.
To do so, we divide the specification in 2 logical parts : data discovery, dealing with metadata (based on SimDM) and data access, dealing with the concrete data (preview, raw data extraction).
Data discovery
Common use case of access to simulation results
The common user information search use cases are:
-
Searches for a single simulation
-
Give me a cube of Universe at redshift 10 simulated with Lambda CDM cosmology
-
Give ma a theoretical stellar spectrum for a B 0 star
-
-
Searches for simulations in a grid of models
-
Give me the density (input parameters) of simulations producing column densities of CO between 1E14 and 4E14 cm-2 (inverse problem)
-
Give me all the simulations in the input parameter range : InputParameter_ 1 between p1 and p2, and InputParameter_2 between q1 and q2
-
Then the SimDAL service should propose to download parts of the corresponding data. The following describes a protocol allowing to run this use cases a standard way.
Discover SimDAL services through registry requests based on SKOS concepts
The first step is to discover relevant SimDAL services through the VO
registries
[For the moment, we think about "registry" in the sense
"IVOA Registries" (not SimDB)]
.
Each SimDAL service is registered in registries with the following minimal informations :
-
a list of SimTAP protocol file (xml) (1 for each protocol on which the simDAL service has results for)
-
a list of the SKOS concepts related to each protocol (even if this information is already in the protocol xml file, it is useful to give them here for fast search)
Give me all the resources
[SimDM meaning]
(and thus the SimDAL
services) dealing with the "c" SKOS concept
http://purl.org/astronomy/vocab/PhysicalProcesses/Magnetohydrodynamics http://purl.org/astronomy/vocab/AstronomicalObjects/Star
Users/clients should be able to discover SimDAL services searching in the
registries
[How to register SimDAL services in VO registries will be
defined later with the Registry W.G.]
on the SKOS concepts describing projects.
SimTAP
SimDAL is a specification for services by which publishers can publish simulation data obtained by running simulation codes. The simulation codes can be described by the simulation data model (SimDM), and can the results of the simulations. SimDM can be mapped to a relational model and a TAP_SCHEMA and therefore a TAP service could be defined over the SimDM to query for interestng simulations, just as ObsTAP is defined over ObsCore.
The main difference between these two examples is that ObsCore is structurally very simple, a single table can store all the ObsCore metadata. In contrast the SimDM is complex, in fact so complex that obvious questions can be asked only with great difficulty in SQL (or ADQL).
We therefore here propose to use an alternative representation of the Simulation Data model for use in the "queryData" part of SimDAL services. This representation allows much simpler queries, but only for a very restricted subset of simulation codes that must be explicitly indicated by the SimDAL service.
Formally, SimTAP is a TAP service on top of a table schema that is constrained by one or more instances of the SimDM:resource/protocol/Protocol class defined in the simulation data model
Here we describe the rules for deriving a SimTAP schema from such SimDM Protocol instances. We there attempt to use UTYPE-s which link to the SimDM html document at [TBD add link], but generally leave out the SimDM:/resource/protocol/ prefix. Note that these rules are implemented in the XSLT script in
-
A SimDAL service SHALL provide information about results of (SimDM:/resource/experiment/)Experiment-s performed according to one or more pre-declared (SimDM:resource/protocol/)Protocol-s.
-
For each Protocol a table is defined. name derived form Protocol.name OR name derived from mapping for Protocol.identity.ivoId.
-
For each Protocol/parameter a column is defined in this table. Name derived from name of parameter OR from entry in mapping file. Datatype derived from parameter datatype OR from mapping file. Values in these columns indicate parameter values for an experiment. I.e. column corresponds to Experiment.parameterValue
-
For each Simulator.physicalProcess a column is defined of type boolean, indicating whether physics has been applied or not. Name of this column is "has"+[name of Physics] OR derived from mapping file. I.e. column corresponds to Experiment.appliedPhysics.
-
For each Protocol.algorithm a column is defined of type boolean, indicating whether algorithm has been applied or not. Name of this column is "has"+[name of Algorithm] OR derived from mapping file. I.e. column corresponds to Experiment.appliedAlgorithm.
-
For each Protocol.outputType an objectType representing the collection of corresponding types. Named [name-of-outputType]+"Collection".
-
for each ObjectType.property the Collection type will get an attribute for one or more statistical summaries. Named after the property.name+"_"+name of statistic. E.g. mass_Nominal.
-
-
For each Protocol.outputType that is also the parent in a composition relationship between different object types we generate a separate class named directly after the object type itself. This class represents individual objects with their properties. In the current implementation [TBD decide on this!] the collection type for each child object type will be given a reference to the parent object. An alternative implementation would have the collection types be contained by the parent type.
-
TODO represent resource metadata into experiment class; represent target(s);
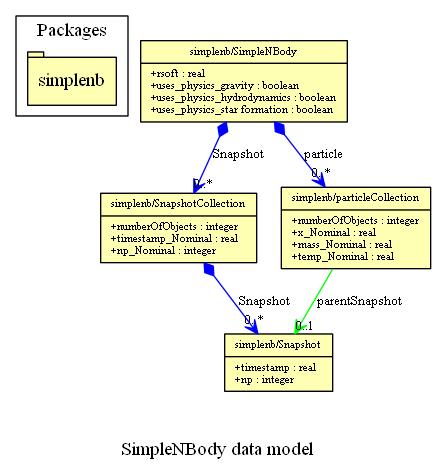
As an example consider the following instance diagram of a simple SimDM:/resource/protocol/Simulator.

The formal SimDM XML representation of that instance diagram is given in Appendix B. Using the mapping rules mentioned above we have written an XSLT script that translates that XML document to a .vo-urp representation, reproduced in Appendix B as well. That .vo-urp document in its right was input to part of the XSLT pipeline from VO-URP to produce a TAP_SCHEMA and a HTML document. The latter contains also a GraphViz generated diagram illustrating the generated model, which is reproduced below. Note that at no point was it necessary to produce a UML model in this process. All results are obtained from the simple SimDM/Protocol document. The VO-URP pipeline was used only for convenience. In principle the TAP schema could have been produced by hand according to a set of mapping rules directly from the SimDM/Protocol model.
Browse the provider’s tables metadata
to be integrated into SimTAP part ?
The idea here is to give to the provider the responsability to provide tables as relevant "logical views" on the results.
Practically, it is a set of SimDM-derived tables provided through an IVOA TAP implementation. The relevant table/column meaning is described in the TAP_SCHEMA using SimDM formalism (utype, units…) and related Theory SKOS concepts.
In this part, the user discovers which tables (if any) and/or columns exist dealing with the information he is interested in (most of the time it is associated to a SKOS concept).
It is necessary to add 2 mandatory TAP_SCHEMA.columns columns to the TAP standard : group and skos and thus define an extension of TAP : SimTAP.
The concept of group is important in the SimDM (property_group etc…) and maybe used in other contexts as well. Some simulations can have a large number of properties or input parameters. Gathering in groups, is mandatory so that client are able to present informations / tables to the end-user in a usable way. In addition, it allows provider to subjectively add new meaning to its data by grouping table columns as well as presentation ease.
Practically, the group concept could be added in TAP_SCHEMA through a special TAP_SCHEMA.groups metadata table.
<?xml version="1.0" encoding="utf-8"?> <VOTABLE xmlns="http://www.ivoa.net/xml/VOTable/v1.2" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" version="1.2" xsi:schemaLocation="http://www.ivoa.net/xml/VOTable/v1.2 http://vo.ari.uni-heidelberg.de/docs/schemata/VOTable-1.2.xsd"> <RESOURCE> <INFO name="server" value="http://localhost:8080"/> <INFO name="query" value="SELECT * FROM tap_schema.groups LIMIT 2000"/> <INFO name="src_res" value="Contains traces from resource __system__/tap">Data and services dealing with the Table Access Protocol TAP.</INFO> <INFO name="src_table" value="Contains traces from table tap_schema.groups">Columns that are part of groups within tables available for ADQL querying.</INFO> <TABLE name="groups"> <FIELD datatype="char" ID="table_name" name="table_name" arraysize="*"> <DESCRIPTION>Fully qualified table name</DESCRIPTION> </FIELD> <FIELD datatype="char" ID="column_name" name="column_name" arraysize="*"> <DESCRIPTION>Name of a column belonging to the group</DESCRIPTION> </FIELD> <FIELD datatype="char" ID="column_utype" name="column_utype" arraysize="*"> <DESCRIPTION>utype the column withing the group</DESCRIPTION> </FIELD> <FIELD datatype="char" ID="group_name" name="group_name" arraysize="*"> <DESCRIPTION>Name of the group</DESCRIPTION> </FIELD> <FIELD datatype="char" ID="group_utype" name="group_utype" arraysize="*"> <DESCRIPTION>utype of the group</DESCRIPTION> </FIELD> <DATA> <TABLEDATA> <TR> <TD>dataset_123_halos2.exts</TD> <TD>r178</TD> <TD>SimDM:/resource/property</TD> <TD>group178</TD> <TD>SimDM:/resource/propertyGroup</TD> </TR> <TR> <TD>dataset_123_halos2.exts</TD> <TD>v178</TD> <TD>SimDM:/resource/property</TD> <TD>group178</TD> <TD>SimDM:/resource/propertyGroup</TD> </TR> </TABLEDATA> </DATA> </TABLE> </RESOURCE> </VOTABLE>
As a general pattern in the VO environment, the data discovery is done through TAP queries on VO datamodel entities (utype, ucd). SimDAL must follow this pattern for its discovery part through SimTAP, adding query capabilities on group and skos.
Browse data table
Once, the tables and/or columns of interest are found, one want to browse them through standard TAP tools (TopCat, tapsh…). Specific simulation search features (like per skos, per group) would be nicely integrated to existing VO tools like TopCat through plugins (in the same way as the integration of BastI).
If a specific presentation is needed, a customized web user interface can be developped on top of an ADQL adapter very easily.
Once data of interest is found
One may look for any information provided by the provider’s tables. However, an experiment satisfying some characteristics is the common searched entity. Once it is done, the user will want to preview, access the corresponding infos (output datasets, code parameter settings…). To do so, he will have to be able to reference this particular entity (simulation for ex).
A possible approach to this requirement would be to identify an identity through a specific ID to be used later in data access functions (preview, cutout…). This ID must carry 2 informations . the service global ID (unique in the world) : 4 char ASCII (like airports codes ?). The one already settled for registry could be used as well (ex: ivo://obspm.fr/organisation) . the element local ID : 16 char ASCII
The ID pattern may be : <GLOBAL_ID>:<LOCAL_ID> ex: LPAR:simu_ram_0000012 (or ivo://obspm.fr/organisation:simu_ram_0000012)
Preview
As often as possible, the provider may provide a subjective preview of his results (simulation as a whole, dataset, subdataset etc…). The preview can be any web downloadable url (http pointing jpeg img, plaintext file, fits etc….) and can represent any SimDM resource. (See reflexion from Laurent Michel (CDS) on scientists needs survey for TAPHandle.)
A simple and straightforward solution would be to standardize a column name preview in SimTAP tables which, when present, would contain an url toward a preview of the current table row. A parallel with IVOA’s datalink could be done to enable multi previews feature (for example a preview url points toward an xml file listing & describing all previews url related to the current row).
This way, preview on a table as a whole and on a column as a whole would be handled by TAP_SCHEMA.{columns,tables} special preview column. For preview on a single row of the data, a normal column preview should be added in the data table.
Semantic
A semantic layer is the key of a powerful, scalable and robust discovery system. The available prototype using SKOS concepts (PoolParty) at VO-Paris shows the power of this approach and must be integrated in the SimDAL core the same way it is in SimDM.
It may be done through a SimTAP’s TAP_SCHEMA standardized keyword skos for both TAP_SCHEMA.columns and TAP_SCHEMA.tables
PQL
PQL could be very interesting to query the SimDM, which can’t be always simplified by derivating pivoted tables (and be queried through TAP).
Moreover, the community needs a prototype implementation to show the potential and feasibility of PQL. It could be the opportunity to do so.
Data Access
formatted dataset (result) vs raw dataset
The raw output of a code run (ex IDL binary files) must be differentiated from formatted subset of it, named Result. The later is designed to focus on the scientific meaning of the experiment result shared by the publisher. The former is just a technical binary file.
Whatever the format is, it must be accessible a standard way.
It may be relevant not to over define this part, letting the provider the
choice to use any url downloadable format like HDF5, VTK, FITS, jpeg, plain
text file
[For tabular data (those obtained through TAP queries)
VOTable is the preferred format.]
. However, a way to inform the user about
which format is used must be defined. This could be done through the VOSI
compliant part of the service.
Usually, data from TAP queries (provider’s results) will be exchanged through VOTable whereas raw data will be in binary form.
cutout on formatted dataset
The need for a way to extract subdatasets from simulations results
[i.e SimDM outputData]
is a core requierement since the beginning of
SimDAL (see Gheller, Lemson, Wagner first SimDAP note). For some time, it has
been becoming a central issue for the traditional DAL WG as well.
The solution design, matured during the past years by the Theory IG members, is a simple per-axis restriction. It is based on a RESTful like url pattern.
cutout: string list list list -> dataset to extract a subdataset of datasetId restricted according to attributes_restriction and where only attributes_list attributes of subdataset's objects are present. Apply provided options. cutout(dataset_id, attribute_list, attribute_restrictions_list, options_list)
original_dataset <- { id:Halo23_ramses_34,
data:
[
{mass:1.23e2, nbr_part:3.45e5, ener_pot:2.01, x:1, y:2,z:0},
{mass:1.03e2, nbr_part:2.89e5, ener_pot:1.71, x:23,y:4,z:4},
{mass:3.673e3, nbr_part:9.45e5, ener_pot:2.41, x:4,y:5,z:3},
{mass:1.2e1, nbr_part:1.45e3, ener_pot:0.81, x:3,y:7,z:3}
]
}
attribute_list <- [mass,nbr_part]
attribute_restriction_list <- [
{attribute : x,condition: gt,restriction:0},
{attribute : x,condition: lt,restriction:15},
{attribute : y,condition: gt,restriction:3},
{attribute : y,condition: lt,restriction:8},
{attribute : z,condition: gt,restriction:2},
{attribute : z,condition: lt,restriction:4},
{attribute : mass, condition: ordered, restriction:asc}
]
cutout(Halo23_ramses_34,attribute_list, attribute_restriction_list) should produce :
[
{mass:1.2e1, nbr_part:1.45e3, ener_pot:0.81, x:3,y:7,z:3},
{mass:3.673e3, nbr_part:9.45e5, ener_pot:2.41, x:4,y:5,z:3}
]
input
- option
-
an options list argument must be supported, mainly to allow user control on the amount of data the service will return. To address that we represent a dataset as an ordered list of pages. Each page is identified by a tuple (offset,limit), to be read page starting at record offset and containing limit records the options list can currently contain:
-
offset: number
-
limit: number (i.e page size)
-
output_format: one of the service supported format (XML/VOTABLE, txt, google protobuf…). Default to VOTABLE.
-
- attributes_list
-
A list of available attributes
[That is those described in the protocol file, i.e objecttype’s properties]
- attributes_restrictions_list
-
A list of restrictions to apply to available properties of objects in the selected dataset. a restriction rule has the following form:
{attribute : string, condition: [gt|lt|ordered], restriction:[string|number|asc|desc]}The condition can be:
-
greater than, gt
-
lower than, lt
-
order by, ordered, in this case, the restriction attribute is asc or desc
-
- dataset_id
-
A string representing the data identifier in the service.
output
The output file produced by the service. It contains the attributes_list properties of the elements of the datasetId dataset verifying the attributes_restrictions_list constraints. The file format is the one provided in the options parameter.
cutout on raw dataset
Same as formatted dataset but with different available output file formats. Since the amount of data can be very large, the service must be based on UWS and in particular support an async mode.
Preview
An important part guiding the user in his datamining task are preview data. A possible way to standardize it is the one previously exposed.
The protocol to access preview material is the HTTP one, there are a lot of light http client implementations in many languages able to retrieve and show standards url refering standard mime-type files.
Furthermore, we can notice that a very useful feature from S3 : summary is easily abstracted by this concept of preview.
Data Link
Make SimDAL able to use DataLink specification could be relevant, in particular to enforce the link simulation data <→ observational data (see François Bonnarel).
The most relevant use is the link toward previews and any static raw binary data. This way we could add the possibility to fill the preview field with a link toward a datalink resource (i.e an xml file listing links toward several previews files).
<?xml version="1.0" encoding="utf-8"?> <DATALINK> <LINK> <URL>http://roxxor.obspm.fr/deuvo-ui/dfiles//simtap.objects_34_halo/votable?select=x%2Cy%2Cz%2Cmass%2Cnpart&where=npart+%3E+2e4</URL> <MIME>application/xml</MIME> <DESCRIPTION>subdataset of the fof halo finder postprocessing on top of a Ratra-Peebles universe simulation (boxlength 162, resolution 1024, z=1.5) output. Constraints are number of particles gt 2e4</DESCRIPTION> <SIZE>unknown</SIZE> </LINK> <LINK> <URL> http://roxxor.obspm.fr/deuvo-ui/dfiles//simtap.objects_32_halo/votable?select=x%2Cy%2Cz%2Cmass%2Cnpart&where=npart+%3E+2e4</URL> <MIME>application/xml</MIME> <DESCRIPTION>subdataset of the fof halo finder postprocessing on top of a Ratra-Peebles universe simulation (boxlength 162, resolution 1024, z=2.33) output. Constraints are number of particles gt 2e4</DESCRIPTION> <SIZE>unknown</SIZE> </LINK> </DATALINK>
Exchange format
TBD
Posponed
Appendix A: Examples of typical SimDAL use case
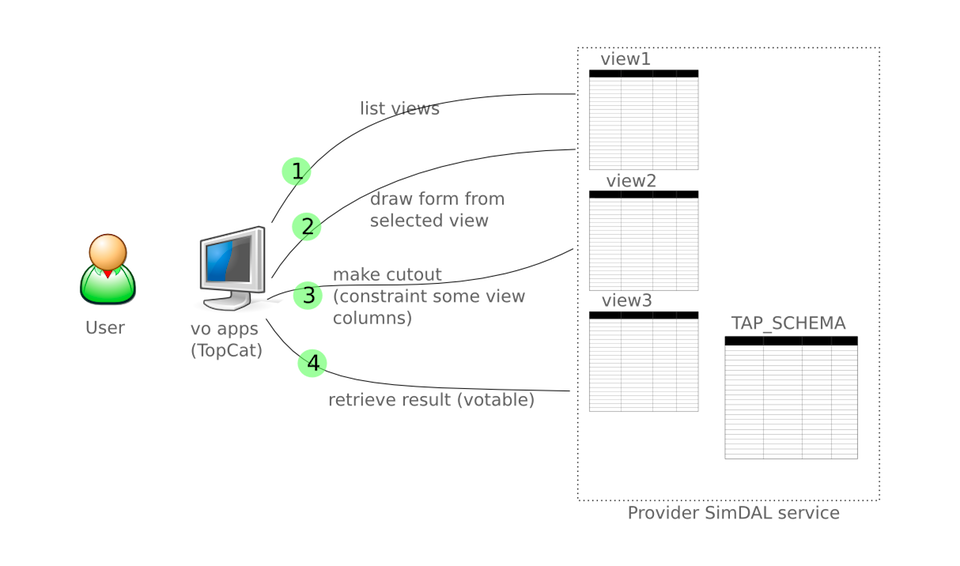
User accessing SimDAL through vo app
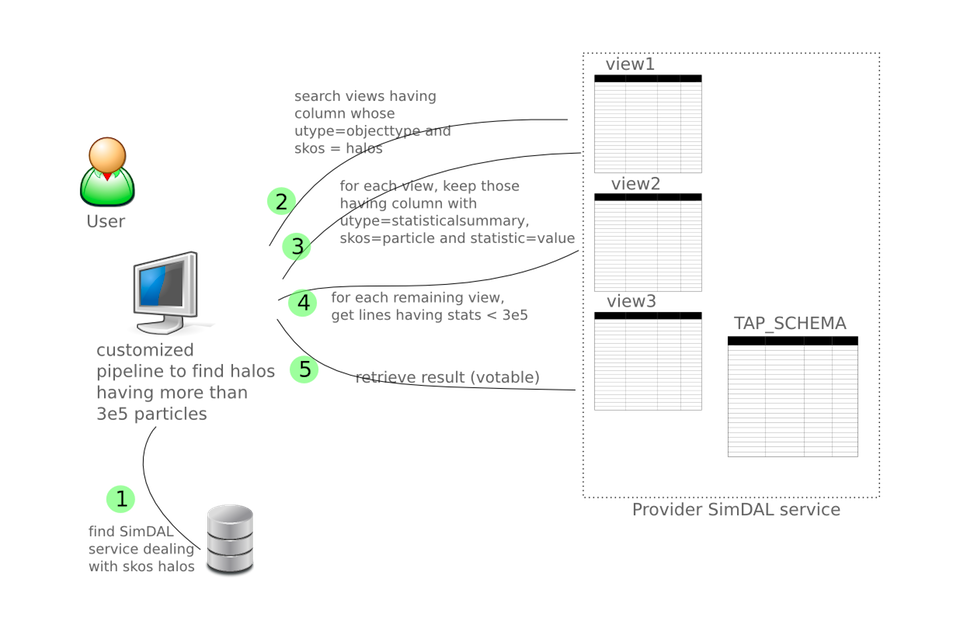
User defining an automated batch pipeline
Appendix B: SimTAP input/output
The SimDM/XML representation of a simple Simulator used in the SimTAP section above.
<?xml version="1.0" encoding="UTF-8" standalone="yes"?> <ns2:aSimulator xmlns:ns2="http://www.ivoa.net/xml/SimDM/v1.0"> <identity publisherDID="ivo://www.mpa-garching.mpg.de/simplenbody"/> <name>SimpleNBody</name> <description>A simple N-Body simulation code.</description> <referenceURL>http://www.mpa-garching.mpg.de/simpleNBody</referenceURL> <created>2009-06-05T14:57:46.047+02:00</created> <updated>2009-06-05T14:57:46.047+02:00</updated> <status>published</status> <contact> <role>owner</role> <party publisherDID="ivo://mpa-garching.mpg.de/lemson"/> </contact> <version>1</version> <!-- algorithm --> <algorithm><name>pp</name></algorithm> <algorithm><name>tree</name></algorithm> <algorithm><name>SPH</name></algorithm> <!-- InputParameter --> <parameter> <identity publisherDID="ivo://www.mpa-garching.mpg.de/simplenbody#param/rsoft" xmlId="PAR_rsoft"/> <name>rsoft</name> <datatype>real</datatype> <cardinality>1</cardinality> <description>The softening radius.</description> </parameter> <outputType> <identity publisherDID="ivo://www.mpa-garching.mpg.de/simpeNBody#Snapshot" /> <name>Snapshot</name> <description>A Snapshot is a representation of the state of the simulation at a given time. It consists of files listing the properties of simulation particles of up to 6 types.</description> <property> <identity publisherDID="ivo://www.mpa-garching.mpg.de/simpeNBody#Snapshot/Timestamp"/> <name>timestamp</name> <datatype>real</datatype> <cardinality>1</cardinality> <description>The timestamp at which the Snapshtot was created.</description> </property> <property> <identity publisherDID="ivo://www.mpa-garching.mpg.de/simpeNBody#Snapshot/Np"/> <name>np</name> <datatype>integer</datatype> <cardinality>1</cardinality> <description>The number of particles in the snpashot</description> </property> <relationship> <identity publisherDID="ivo://www.mpa-garching.mpg.de/simpeNBody#Snapshot/Particles"/> <name>Particles</name> <description>Collection of particles in a snapshot.</description> <relationshipType>composition</relationshipType> <cardinality>0..*</cardinality> <relatedObjectType xmlId="Particle"/> </relationship> </outputType> <outputType> <identity xmlId="Particle" publisherDID="ivo://www.mpa-garching.mpg.de/simpeNBody#Particle" /> <name>particle</name> <description>TBD</description> <property> <identity publisherDID="ivo://www.mpa-garching.mpg.de/simpeNBody#Particle/X" xmlId="Particle_X"/> <name>x</name> <datatype>real</datatype> <cardinality>1</cardinality> <description>X (cartesian) coordinate of the particle.</description> <label>http://purl.org/astronomy/vocab/PhysicalQuantities/PositionOfMassCentroid#X</label> </property> <property> <identity publisherDID="ivo://www.mpa-garching.mpg.de/simpeNBody#Particle/Mass" xmlId="Mass"/> <name>mass</name> <datatype>real</datatype> <cardinality>1</cardinality> <description>Mass of particle.</description> <label>http://purl.org/astronomy/vocab/PhysicalQuantities/Mass</label> </property> <property> <identity publisherDID="ivo://www.mpa-garching.mpg.de/simpeNBody#Particle/Temp" xmlId="Temp"/> <name>temp</name> <datatype>real</datatype> <cardinality>0..1</cardinality> <description>Temperature of particle.</description> </property> <property> <identity publisherDID="ivo://www.mpa-garching.mpg.de/simpeNBody#Particle/ParticleType" xmlId="ParticleType"/> <name>ParticleType</name> <datatype>string</datatype> <cardinality>1</cardinality> <description>Type of particle.</description> <isEnumerated>true</isEnumerated> <validValue><value>gas</value></validValue> <validValue><value>star</value></validValue> <validValue><value>dm</value></validValue> </property> </outputType> <physicalProcess><name>gravity</name><description>Gravitational interaction between particles.</description> <label>http://purl.org/astronomy/vocab/PhysicalProcesses/Gravitation</label> </physicalProcess> <physicalProcess><name>hydrodynamics</name><description>Hydrodynamics of gas particles.</description> <label>http://purl.org/astronomy/vocab/PhysicalProcesses/Hydrodynamics</label> </physicalProcess> <physicalProcess><name>star formation</name><description>Formation of stars from gas particles.</description> <label>http://purl.org/astronomy/vocab/PhysicalProcesses/StarFormation</label> </physicalProcess> </ns2:aSimulator>
The .vo-urp representation of the SimTAP model generated from the Simulator XML above.
<?xml version="1.0" encoding="UTF-8"?> <model xmiid="MODEL_SimpleNBody"> <name>SimpleNBody</name> <description> A SimTAP model based on the SimDM/Protocol TODO add link to actual model file. </description> <utype>SimpleNBody</utype> <profile xmiid="_10_0_42c01ac_1131096191234_590105_2"> ... </profile> <package xmiid="PACKAGE_simplenb"> <name>simplenb</name> <description>Package containing object types for the SimDM-Protocol SimpleNBody.</description> <utype>SimpleNBody:simplenb/</utype> <objectType xmiid="EXPERIMENT_simplenb_SimpleNBody"> <name>SimpleNBody</name> <utype otherutype="SimDM:resource/experiment/Simulation">SimpleNBody:simplenb/SimpleNBody</utype> <attribute xmiid="EXPERIMENT_PARAMETERVALUE_simplenb_SimpleNBody_rsoft"> <name>rsoft</name> <description> Value of input parameter [rsoft]: The softening radius.</description> <utype otherutype="SimDM:resource/experiment/ParameterValue.numericValue.value">SimpleNBody:simplenb/SimpleNBody.rsoft</utype> <datatype xmiidref="PRIMITIVETYPE_real" name="real"/> <multiplicity>1</multiplicity> </attribute> <attribute xmiid="EXPERIMENT_APPLIEDPHSYICS_simplenb_SimpleNBody_gravity"> <name>uses_physics_gravity</name> <description> Indicates the usage of the physical process gravity: Gravitational interaction between particles.</description> <datatype name="boolean" xmiidref="PRIMITIVETYPE_boolean"/> <multiplicity>0..1</multiplicity> </attribute> <attribute xmiid="EXPERIMENT_APPLIEDPHSYICS_simplenb_SimpleNBody_hydrodynamics"> <name>uses_physics_hydrodynamics</name> <description> Indicates the usage of the physical process hydrodynamics: Hydrodynamics of gas particles.</description> <datatype name="boolean" xmiidref="PRIMITIVETYPE_boolean"/> <multiplicity>0..1</multiplicity> </attribute> <attribute xmiid="EXPERIMENT_APPLIEDPHSYICS_simplenb_SimpleNBody_star formation"> <name>uses_physics_star formation</name> <description> Indicates the usage of the physical process star formation: Formation of stars from gas particles.</description> <datatype name="boolean" xmiidref="PRIMITIVETYPE_boolean"/> <multiplicity>0..1</multiplicity> </attribute> <collection xmiid="EXPERIMENT_OUTPUTTYPE_COLLECTION_simplenb_SimpleNBody_Snapshot"> <name>Snapshot</name> <utype otherutype="SimDM:resource/protocol/Protocol.outputType">SimpleNBody:simplenb/SimpleNBody.Snapshot</utype> <datatype name="SnapshotCollection" xmiidref="EXPERIMENT_OUTPUTDATASET_simplenb_Snapshot"/> <multiplicity>0..*</multiplicity> </collection> <collection xmiid="EXPERIMENT_OUTPUTTYPE_COLLECTION_simplenb_SimpleNBody_particle"> <name>particle</name> <utype otherutype="SimDM:resource/protocol/Protocol.outputType">SimpleNBody:simplenb/SimpleNBody.particle</utype> <datatype name="particleCollection" xmiidref="EXPERIMENT_OUTPUTDATASET_simplenb_particle"/> <multiplicity>0..*</multiplicity> </collection> </objectType> <objectType xmiid="EXPERIMENT_OUTPUTDATASET_simplenb_Snapshot"> <container name="SimpleNBody" xmiidref="EXPERIMENT_simplenb_SimpleNBody" relation="Snapshot"/> <name>SnapshotCollection</name> <description>A Snapshot is a representation of the state of the simulation at a given time. It consists of files listing the properties of simulation particles of up to 6 types.</description> <utype otherutype="SimDM:resource/experiment/Experiment.outputDataSet">SimpleNBody:simplenb/SnapshotCollection</utype> <attribute> <name>numberOfObjects</name> <description>Number of objects in the ouput data set.</description> <datatype name="integer" xmiidref="PRIMITIVETYPE_integer"/> <multiplicity>0..1</multiplicity> </attribute> <attribute xmiid="EXEPRIMENT_OUTPUTDATASET_CHARACTERISATION_simplenb_SnapshotCollection_timestamp_Nominal"> <name>timestamp_Nominal</name> <utype otherutype="SimDM:resource/experiment/StatisticalSummary.numericValue.value">SimpleNBody:simplenb/SnapshotCollection.timestamp_Nominal</utype> <description>Nominal value of property timestamp: The timestamp at which the Snapshtot was created.</description> <datatype name="real" xmiidref="PRIMITIVETYPE_real"/> <multiplicity>0..1</multiplicity> </attribute> <attribute xmiid="EXEPRIMENT_OUTPUTDATASET_CHARACTERISATION_simplenb_SnapshotCollection_np_Nominal"> <name>np_Nominal</name> <utype otherutype="SimDM:resource/experiment/StatisticalSummary.numericValue.value">SimpleNBody:simplenb/SnapshotCollection.np_Nominal</utype> <description>Nominal value of property np: The number of particles in the snpashot</description> <datatype name="integer" xmiidref="PRIMITIVETYPE_integer"/> <multiplicity>0..1</multiplicity> </attribute> <collection xmiid="EXPERIMENT_OUTPUTDATASET_DATAOBJECT_COLLECTION_simplenb_SnapshotCollection_Snapshot"> <name>Snapshot</name> <utype otherutype="SimDM:resource/OutputDataSet.object">SimpleNBody:simplenb/SnapshotCollection.Snapshot</utype> <datatype name="Snapshot" xmiidref="EXPERIMENT_OUTPUTDATASET_DATAOBJECT_simplenb_Snapshot"/> <multiplicity>0..*</multiplicity> </collection> </objectType> <objectType xmiid="EXPERIMENT_OUTPUTDATASET_simplenb_particle"> <container name="SimpleNBody" xmiidref="EXPERIMENT_simplenb_SimpleNBody" relation="particle"/> <name>particleCollection</name> <description>TBD</description> <utype otherutype="SimDM:resource/experiment/Experiment.outputDataSet">SimpleNBody:simplenb/particleCollection</utype> <attribute> <name>numberOfObjects</name> <description>Number of objects in the ouput data set.</description> <datatype name="integer" xmiidref="PRIMITIVETYPE_integer"/> <multiplicity>0..1</multiplicity> </attribute> <attribute xmiid="EXEPRIMENT_OUTPUTDATASET_CHARACTERISATION_simplenb_particleCollection_x_Nominal"> <name>x_Nominal</name> <utype otherutype="SimDM:resource/experiment/StatisticalSummary.numericValue.value">SimpleNBody:simplenb/particleCollection.x_Nominal</utype> <description>Nominal value of property x: X (cartesian) coordinate of the particle.</description> <datatype name="real" xmiidref="PRIMITIVETYPE_real"/> <multiplicity>0..1</multiplicity> </attribute> <attribute xmiid="EXEPRIMENT_OUTPUTDATASET_CHARACTERISATION_simplenb_particleCollection_mass_Nominal"> <name>mass_Nominal</name> <utype otherutype="SimDM:resource/experiment/StatisticalSummary.numericValue.value">SimpleNBody:simplenb/particleCollection.mass_Nominal</utype> <description>Nominal value of property mass: Mass of particle.</description> <datatype name="real" xmiidref="PRIMITIVETYPE_real"/> <multiplicity>0..1</multiplicity> </attribute> <attribute xmiid="EXEPRIMENT_OUTPUTDATASET_CHARACTERISATION_simplenb_particleCollection_temp_Nominal"> <name>temp_Nominal</name> <utype otherutype="SimDM:resource/experiment/StatisticalSummary.numericValue.value">SimpleNBody:simplenb/particleCollection.temp_Nominal</utype> <description>Nominal value of property temp: Temperature of particle.</description> <datatype name="real" xmiidref="PRIMITIVETYPE_real"/> <multiplicity>0..1</multiplicity> </attribute> <reference> <name>parentSnapshot</name> <utype>SimpleNBody:simplenb/particleCollection.parentSnapshot</utype> <datatype xmiidref="EXPERIMENT_OUTPUTDATASET_DATAOBJECT_simplenb_Snapshot" name="Snapshot"/> <multiplicity>0..1</multiplicity> </reference> </objectType> <objectType xmiid="EXPERIMENT_OUTPUTDATASET_DATAOBJECT_simplenb_Snapshot"> <container name="SimpleNBody" xmiidref="EXPERIMENT_simplenb_SimpleNBody" relation="Snapshot"/> <name>Snapshot</name> <description>A Snapshot is a representation of the state of the simulation at a given time. It consists of files listing the properties of simulation particles of up to 6 types.</description> <utype otherutype="SimDM:resource/experiment/DataObject">SimpleNBody:simplenb/Snapshot</utype> <attribute xmiid="EXEPRIMENT_OUTPUTDATASET_DATAOBJECT_PROPERTYVALUE_simplenb_Snapshot_timestamp"> <name>timestamp</name> <utype otherutype="SimDM:resource/experiment/PropertyValue.numericValue.value">SimpleNBody:simplenb/Snapshot.timestamp</utype> <description> Value of property timestamp: The timestamp at which the Snapshtot was created.</description> <datatype name="real" xmiidref="PRIMITIVETYPE_real"/> </attribute> <attribute xmiid="EXEPRIMENT_OUTPUTDATASET_DATAOBJECT_PROPERTYVALUE_simplenb_Snapshot_np"> <name>np</name> <utype otherutype="SimDM:resource/experiment/PropertyValue.numericValue.value">SimpleNBody:simplenb/Snapshot.np</utype> <description> Value of property np: The number of particles in the snpashot</description> <datatype name="integer" xmiidref="PRIMITIVETYPE_integer"/> </attribute> </objectType> </package> </model>
The TAP representation of the SimTAP model generated from the Simulator XML above. The schema is represented as <TABLE>-s in a VOTable.
<?xml version="1.0" encoding="UTF-8"?> <VOTABLE xlmns="http://www.ivoa.net/xml/VOTable/v1.1" version="1.1"> <RESOURCE name="SimpleNBody"> <DESCRIPTION> A SimTAP model based on the SimDM/Protocol TODO add link to actual model file. </DESCRIPTION> <RESOURCE name="simplenb"> <DESCRIPTION>Package containing object types for the SimDM-Protocol SimpleNBody.</DESCRIPTION> <TABLE name="SimpleNBody"> <DESCRIPTION/> <FIELD name="ID" datatype="long" ucd="TBD" utype="SimpleNBodySimpleNBody:simplenb/SimpleNBodyrsoft
				Value of input parameter [rsoft]:
				The softening radius.SimpleNBody:simplenb/SimpleNBody.rsoft1uses_physics_gravity
				Indicates the usage of the physical process	gravity:
				Gravitational interaction between particles.0..1uses_physics_hydrodynamics
				Indicates the usage of the physical process	hydrodynamics:
				Hydrodynamics of gas particles.0..1uses_physics_star formation
				Indicates the usage of the physical process	star formation:
				Formation of stars from gas particles.0..1SnapshotSimpleNBody:simplenb/SimpleNBody.Snapshot0..*particleSimpleNBody:simplenb/SimpleNBody.particle0..*"> <DESCRIPTION>The unique, primary key column on this table.</DESCRIPTION> </FIELD> <FIELD name="DTYPE" datatype="char" width="32" ucd="TBD" utype="TBD"> <DESCRIPTION>This column stores the name of the object type from the data model stored in the row.</DESCRIPTION> </FIELD> <FIELD name="rsoft" datatype="double" utype="SimpleNBody:simplenb/SimpleNBody.rsoft"> <DESCRIPTION> Value of input parameter [rsoft]: The softening radius.</DESCRIPTION> </FIELD> <FIELD name="uses_physics_gravity" datatype="boolean" utype="SimpleNBody:simplenb/SimpleNBody.uses_physics_gravity"> <DESCRIPTION> Indicates the usage of the physical process gravity: Gravitational interaction between particles.</DESCRIPTION> </FIELD> <FIELD name="uses_physics_hydrodynamics" datatype="boolean" utype="SimpleNBody:simplenb/SimpleNBody.uses_physics_hydrodynamics"> <DESCRIPTION> Indicates the usage of the physical process hydrodynamics: Hydrodynamics of gas particles.</DESCRIPTION> </FIELD> <FIELD name="uses_physics_star formation" datatype="boolean" utype="SimpleNBody:simplenb/SimpleNBody.uses_physics_star formation"> <DESCRIPTION> Indicates the usage of the physical process star formation: Formation of stars from gas particles.</DESCRIPTION> </FIELD> </TABLE> <TABLE name="Snapshot"> <DESCRIPTION>A Snapshot is a representation of the state of the simulation at a given time. It consists of files listing the properties of simulation particles of up to 6 types.</DESCRIPTION> <FIELD name="ID" datatype="long" ucd="TBD" utype="SnapshotA Snapshot is a representation of the state of the simulation at a given time. 
 It consists of files listing the properties of simulation particles of up to 6 types.SimpleNBody:simplenb/SnapshottimestampSimpleNBody:simplenb/Snapshot.timestamp
				Value of property timestamp:
				The timestamp at which the Snapshtot was created.npSimpleNBody:simplenb/Snapshot.np
				Value of property np:
				The number of particles in the snpashot"> <DESCRIPTION>The unique, primary key column on this table.</DESCRIPTION> </FIELD> <FIELD name="DTYPE" datatype="char" width="32" ucd="TBD" utype="TBD"> <DESCRIPTION>This column stores the name of the object type from the data model stored in the row.</DESCRIPTION> </FIELD> <FIELD name="containerId" datatype="long" ucd="TBD" utype="SimpleNBody:simplenb/Snapshot.CONTAINER"> <DESCRIPTION>This column is a foreign key pointing to the containing object in SimpleNBody</DESCRIPTION> </FIELD> <FIELD name="timestamp" datatype="double" utype="SimpleNBody:simplenb/Snapshot.timestamp"> <DESCRIPTION> Value of property timestamp: The timestamp at which the Snapshtot was created.</DESCRIPTION> </FIELD> <FIELD name="np" datatype="integer" utype="SimpleNBody:simplenb/Snapshot.np"> <DESCRIPTION> Value of property np: The number of particles in the snpashot</DESCRIPTION> </FIELD> </TABLE> <TABLE name="SnapshotCollection"> <DESCRIPTION>A Snapshot is a representation of the state of the simulation at a given time. It consists of files listing the properties of simulation particles of up to 6 types.</DESCRIPTION> <FIELD name="ID" datatype="long" ucd="TBD" utype="SnapshotCollectionA Snapshot is a representation of the state of the simulation at a given time. 
 It consists of files listing the properties of simulation particles of up to 6 types.SimpleNBody:simplenb/SnapshotCollectionnumberOfObjectsNumber of objects in the ouput data set.0..1timestamp_NominalSimpleNBody:simplenb/SnapshotCollection.timestamp_NominalNominal value of property timestamp:
				The timestamp at which the Snapshtot was created.0..1np_NominalSimpleNBody:simplenb/SnapshotCollection.np_NominalNominal value of property np:
				The number of particles in the snpashot0..1SnapshotSimpleNBody:simplenb/SnapshotCollection.Snapshot0..*"> <DESCRIPTION>The unique, primary key column on this table.</DESCRIPTION> </FIELD> <FIELD name="DTYPE" datatype="char" width="32" ucd="TBD" utype="TBD"> <DESCRIPTION>This column stores the name of the object type from the data model stored in the row.</DESCRIPTION> </FIELD> <FIELD name="containerId" datatype="long" ucd="TBD" utype="SimpleNBody:simplenb/SnapshotCollection.CONTAINER"> <DESCRIPTION>This column is a foreign key pointing to the containing object in SimpleNBody</DESCRIPTION> </FIELD> <FIELD name="numberOfObjects" datatype="integer" utype="SimpleNBody:simplenb/SnapshotCollection.numberOfObjects"> <DESCRIPTION>Number of objects in the ouput data set.</DESCRIPTION> </FIELD> <FIELD name="timestamp_Nominal" datatype="double" utype="SimpleNBody:simplenb/SnapshotCollection.timestamp_Nominal"> <DESCRIPTION>Nominal value of property timestamp: The timestamp at which the Snapshtot was created.</DESCRIPTION> </FIELD> <FIELD name="np_Nominal" datatype="integer" utype="SimpleNBody:simplenb/SnapshotCollection.np_Nominal"> <DESCRIPTION>Nominal value of property np: The number of particles in the snpashot</DESCRIPTION> </FIELD> </TABLE> <TABLE name="particleCollection"> <DESCRIPTION>TBD</DESCRIPTION> <FIELD name="ID" datatype="long" ucd="TBD" utype="particleCollectionTBDSimpleNBody:simplenb/particleCollectionnumberOfObjectsNumber of objects in the ouput data set.0..1x_NominalSimpleNBody:simplenb/particleCollection.x_NominalNominal value of property x:
				X (cartesian) coordinate of the particle.0..1mass_NominalSimpleNBody:simplenb/particleCollection.mass_NominalNominal value of property mass:
				Mass of particle.0..1temp_NominalSimpleNBody:simplenb/particleCollection.temp_NominalNominal value of property temp:
				Temperature of particle.0..1parentSnapshotSimpleNBody:simplenb/particleCollection.parentSnapshot0..1"> <DESCRIPTION>The unique, primary key column on this table.</DESCRIPTION> </FIELD> <FIELD name="DTYPE" datatype="char" width="32" ucd="TBD" utype="TBD"> <DESCRIPTION>This column stores the name of the object type from the data model stored in the row.</DESCRIPTION> </FIELD> <FIELD name="containerId" datatype="long" ucd="TBD" utype="SimpleNBody:simplenb/particleCollection.CONTAINER"> <DESCRIPTION>This column is a foreign key pointing to the containing object in SimpleNBody</DESCRIPTION> </FIELD> <FIELD name="numberOfObjects" datatype="integer" utype="SimpleNBody:simplenb/particleCollection.numberOfObjects"> <DESCRIPTION>Number of objects in the ouput data set.</DESCRIPTION> </FIELD> <FIELD name="x_Nominal" datatype="double" utype="SimpleNBody:simplenb/particleCollection.x_Nominal"> <DESCRIPTION>Nominal value of property x: X (cartesian) coordinate of the particle.</DESCRIPTION> </FIELD> <FIELD name="mass_Nominal" datatype="double" utype="SimpleNBody:simplenb/particleCollection.mass_Nominal"> <DESCRIPTION>Nominal value of property mass: Mass of particle.</DESCRIPTION> </FIELD> <FIELD name="temp_Nominal" datatype="double" utype="SimpleNBody:simplenb/particleCollection.temp_Nominal"> <DESCRIPTION>Nominal value of property temp: Temperature of particle.</DESCRIPTION> </FIELD> <FIELD name="parentSnapshotId" datatype="long" ucd="TBD" utype="TBD"> <DESCRIPTION> This column is a foreign key pointing to the referenced object in Snapshot. </DESCRIPTION> </FIELD> </TABLE> </RESOURCE> </RESOURCE> </VOTABLE>